AlphaFold 3
Written and illustrated by Laurianne Daoust.
Prefer video content? Episode 1 of our Lab Rats series on AlphaFold 3 can be found here.
AlphaFold 2 — A brief history
“Proteins: they are the molecular machines of life!” said each of my life science teachers. The building blocks of these machines, amino acids, are assembled in every cell to create complex 3D structures. Since a protein’s structure dictates its function, scientists have long been evaluating protein shape through methods such as X-ray crystallography or cryo–electron microscopy. With these experimental techniques, the structure of 170,000 proteins has been determined—however, this is a small amount when compared to the 200 million total known proteins [1]. Needless to say, progress was slow, and the following question arose: how can you determine a protein’s structure solely from its amino acid sequence? This decades-old question is known as the Protein Folding Problem—and some believe that AlphaFold 2 had solved it [2].
Unfolded proteins are like a necklace of beads (i.e., amino acids). The interactions between the amino acids on the necklace determine the folding of the protein, but given the abundance of amino acids within each protein and the multitude of potential interactions between each pair, there are millions of possible structures for each protein [1]. AlphaFold 2.0—a deep learning-based system developed by DeepMind and Isomorphic Labs—was designed to predict the 3D structure of a protein from its amino acid sequence. AlphaFold uses a type of advanced computer program called deep learning, which combines two techniques: convolutional neural networks (CNNs), which are good at recognizing patterns in data, and attention mechanisms, which help the program focus on the most important parts of the data [3].
AlphaFold 2 — How it works
To determine the correct protein structure, scientists look at many possible shapes and calculate how much free energy (i.e., energy available to do useful work) each one has. Proteins that aren't folded have high free energy and entropy (i.e., randomness), while folded proteins have low free energy and are more orderly, and thus stable [4]. Determining the correct protein structure is a major challenge due to the astronomical number of possible structures [1, 4].
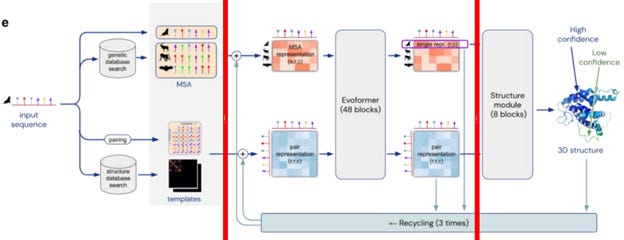
AlphaFold 2 helps by taking the sequence of amino acids and comparing it to sequences of known proteins [3, 5]. This comparison, known as multiple sequence alignment (MSA), identifies proteins with similar structures to the template inputs and creates a pair representation showing how all pairs of amino acids in the target protein interact (see Figure 1).
Then, AlphaFold 2 uses a tool called a transformer (known as Evoformer in AlphaFold 2 or Pairformer in AlphaFold 3) to refine these representations by exchanging information between the MSA and the pair interactions. Finally, this information reaches the structure module, which uses a deep learning model to predict the 3D coordinates of the atoms in the protein in a single step [3, 5].
Figure 1. Flow diagram of the functioning of AlphaFold 2. AlphaFold 3 uses Pairformer (and a Diffusion module) rather than Evoformer as a transformer. Adapted from [5].
AlphaFold 3 - How It Works and How to Use it
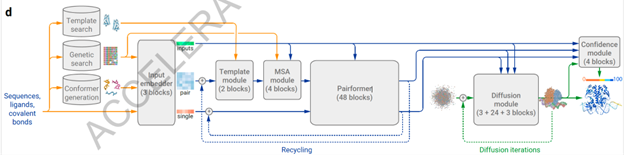
Figure 2. Flow diagram of the functioning of AlphaFold 3. [6]
Essentially, the MSA sequences and templates pass through a simpler version of the previous Evoformer transformer in AlphaFold 3: Pairformer. This transformer’s raw predictions are then fed to a Diffusion module, which begins with a cloud of atoms placed at random and uses these predictions to iteratively mold that cloud into an accurate representation of the input sequences’ 3D structure [7].
AlphaFold 3 is accessible for free here: alphafoldserver.com
AlphaFold 3 - What’s new and groundbreaking
Increased accuracy: AlphaFold 3’s protein structure prediction accuracy for antibodies has more than doubled when compared to AlphaFold 2.3.
Broader molecular predictions: Unlike its predecessor, AlphaFold 3 has been enhanced to predict the structures of not only proteins, but also DNA, RNA, ions, modified residues, and other small molecules essential for a myriad of biological processes [9]. Therefore, AlphaFold 3 offers higher accuracy for interactions between proteins and other molecules, such as protein-ligand (i.e., small molecules that bind to proteins), protein-nucleic acid (i.e., proteins interacting with DNA or RNA), and antibody-antigen (i.e., antibodies binding to foreign substances) interactions [5]. These improved predictions will create unprecedented capabilities for drug design, as ligands and antibodies are among the most commonly developed drugs!
AlphaFold 3 - Limitations
AlphaFold 3 only captures the static, unchanging structure of molecules. Therefore, dynamic behaviors that often characterize the interaction between a protein and its ligand cannot be modeled by AlphaFold 3 [5].
Since the Diffusion model starts with a randomly generated atom cloud, different runs of AlphaFold 3 might provide different predicted 3D structures, some of which are more or less accurate. To mitigate this issue, you should run AlphaFold 3 a few times to improve accuracy [6].
Main Applications
AlphaFold finds itself relevant in numerous applications, including drug discovery, biotechnology, protein engineering, and vaccine development [8]. AlphaFold's accurate prediction of protein structures allows researchers to better understand the shapes of proteins involved in diseases and aids in the design of drugs that can precisely target these proteins. In particular, AlphaFold 3 offers valuable insights into protein-ligand and protein-protein interactions, an essential aspect in grasping how potential drugs function, as well as in identifying potential drug binding sites [5]. AlphaFold can also be used to design proteins with specific functions, such as enzymes for industrial processes or proteins for therapeutic purposes. By predicting the structures of engineered proteins, researchers can optimize their properties for various applications, such as in the design of enzymes that digest plastic in the oceans [8]. Finally, understanding the structures of viral proteins, such as those involved in viral entry or immune evasion, is crucial for designing vaccines. AlphaFold's predictions can aid in vaccine development by providing insights into the shapes of viral antigens and potential vaccine targets [8].
With AlphaFold 3, it seems that we stand on the brink of a new era in biology, where the secrets of proteins are no longer beyond our grasp; they are just at our fingertips.
References
R. F. Service, “‘The game has changed’: AI triumphs at solving protein structures,” Science, Nov. 2020. [Online]. Available: https://www.science.org/content/article/game-has-changed-ai-triumphs-solving-protein-structures.
A. Al-Janabi, “Has DeepMind’s AlphaFold Solved the Protein Folding Problem?” BioTechniques, vol. 72, no. 3, pp. 73-76, 2022. [Online]. Available: https://www.tandfonline.com/doi/full/10.2144/btn-2022-0007.
C. O. Rubiera, “AlphaFold 2 is here: what’s behind the structure prediction miracle,” Oxford Protein Informatics Group, Jul. 2021. [Online]. Available: https://www.blopig.com/blog/2021/07/alphafold-2-is-here-whats-behind-the-structure-prediction-miracle/.
K. A. Dill, S. B. Ozkan, M. S. Shell, and T. R. Weikl, “The Protein Folding Problem,” Annu. Rev. Biophys., vol. 37, pp. 289-316, 2008. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2443096/.
J. Jumper et al., “Highly accurate protein structure prediction with AlphaFold,” Nature, vol. 596, pp. 583-589, 2021. [Online]. Available: https://www.nature.com/articles/s41586-021-03819-2.
J. Abramson et al., “Accurate structure prediction of biomolecular interactions with AlphaFold 3,” Nature, vol. 630, pp. 493-500, 2024. [Online]. Available: https://www.nature.com/articles/s41586-024-07487-w.
D. Lowe, “AlphaFold 3 Debuts,” Science, May 2024. [Online]. Available: https://www.science.org/content/blog-post/alphafold-3-debuts.
F. Preetham, “AlphaFold 3: A Leap Forward in Biomolecular Structure Prediction — Opportunities and Limitations,” Medium, May 2024. [Online]. Available: https://medium.com/meta-multiomics/alphafold-3-a-leap-forward-in-biomolecular-structure-prediction-opportunities-and-limitations-924350af1181.
Z. Yang, X. Zeng, Y. Zhao, and R. Chen, “AlphaFold2 and its applications in the fields of biology and medicine,” Signal Transduction and Targeted Therapy, vol. 8, no. 115, 2023. [Online]. Available: https://www.nature.com/articles/s41392-023-01381-z.